Phosphoproteins: Where’s the “ON” Switch?
by
 Light switches on the wall: Concepts we take for granted
Light switches on the wall: Concepts we take for granted
For much of my life, dealing with electrical lighting has been relatively straight-forward. Upon walking into a dark room, I flip a wall-mounted switch into the up position to turn the lights on. When I’m done in that room, moving that same switch into the down position turns the lights off. For the first 26 years of my life, turning something on or off had essentially become a “dogma” for me: up for on, down for off.
When I began working as a postdoc at Penn State, I moved into an apartment that had a few unique quirks. One of those was that a lot of the light switches moved in a horizontal direction (left or right) as opposed to almost all vertical switches that I had encountered in my life until then (up or down). Suddenly, “up” no longer necessarily meant “on”, and “down” no longer necessarily meant “off.” The dogma that I had memorized for 26 years prior had been broken!
Phosphoproteins: the light switch is always up-and-down… right? Wrong!
One of the questions I’ve encountered many times in my scientific career- usually coming from students- is how protein phosphorylation affects enzyme activity. Is phosphorylation or dephosphorylation what activates an enzyme? Or, in other words, is the “on” form of an enzyme the phosphorylated or unphosphorylated form, and is the opposite the “off” form? When someone asks me this, student or otherwise, I smile, take a deep breath (mostly to prepare for the onslaught of follow-up questions), and say that it depends entirely on the protein being phosphorylated. Moreover, it can even depend on the phosphorylation site, too; a phosphorylation of a serine residue over here can turn an enzyme on, whereas phosphorylation of a threonine right over there will turn the very same enzyme off. Just like that light switch, which I once took as being “dogma,” sometimes there is no universal answer that is guaranteed to cover every situation.
Mixing and matching: Pathways where phosphorylation is both activating and inactivating
I recently worked on a study which was particularly interesting in terms of phosphorylation activating or inactivating enzymes. This particular signalling pathway contained several protein kinases, some of which were activated by phosphorylation, and others which were inactivated, leading down to the final metabolic target of glycogen synthase (GS). Oh, and, insert shameless plug for my recent publication here.
Most scientists who have taken a biochemistry or molecular/cellular physiology course at the 4th year level have at least a passing familiarity with the insulin signalling pathway. Insulin causes the insulin receptor to phosphorylate and activate the insulin receptor substrate-1 (IRS1). IRS1 interacts with and activates phosphoinositide 3-kinases (PI3Ks), which phosphorylate phosphatidylinositol phosphates (PIPs; for example, converting PIP2 to PIP3). PIP3 recruits both phosphoinositide-dependent kinase-1 (PDK1) and Akt to the plasma membrane, allowing PDK1 to phosphorylate and activate Akt. The take home message, for the scope of this article, is that all of the aforementioned systematic phosphorylations- insulin receptor through Akt, inclusive- are activating. I’d draw you a diagram but when it comes to something so widespread like insulin signalling, I can’t compete with the commercial websites, like this one.
What happens after Akt gets to be even more interesting.
Opposite side of the same coin
Akt is turned on when it is phosphorylated by PDK1. However, Akt is now going to use the same trick- phosphorylation- to turn something off…
Only one protein kinase remains in the signalling pathway originating at the insulin receptor and ending with GS: glycogen synthase kinase-3 (GSK3). However, unlike all the kinases described thus far, GSK3 is actually inactivated by Akt phosphorylation; phosphorylation turns GSK3 OFF. Once inactivated, GSK3 cannot, in turn, phosphorylate its own downstream targets. That means that GS remains unphosphorylated. Like GSK-3, an unphosphorylated GS is an active GS; a phosphorylated GS is an inactive GS.
With some phosphorylations being activating and others being inactivating, while writing the manuscript for that publication I shamelessly plugged, even I admittedly had to keep a few post-it notes glued to my wall so that I could remember which phosphorylation patterns led to an activation or inactivation of the final metabolic target. For your benefit, dear readers, in short, here’s what can happen:
1. Akt active, GSK3 inactive, GS active
When Akt has been phosphorylated and activated by its upstream signalling pathway, it phosphorylates and inactivates GSK3. A phosphorylated GSK3 is inactivated. An inactivated GSK3 cannot phosphorylate GS, and therefore GS remains active. Take a look at Fig 1.

2. Akt inactive, GSK3 active, GS inactive
Now assume Akt has not been phosphorylated by its upstream signalling pathway. An unphosphorylated Akt is inactive. It cannot phosphorylate its downstream targets. Unphosphorylated GSK3 remains active. An active GSK3 phosphorylates GS, inactivating it. Check out Fig. 2.
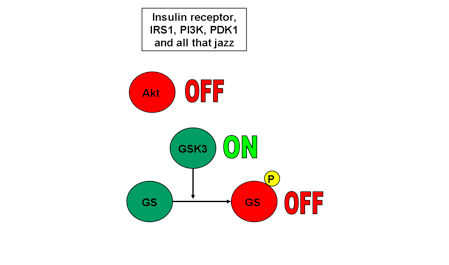
So the next time a student asks me “Chris, does phosphorylation turn an enzyme on or off?” I’ll refer them to this article. And now, you can do the same if a student asks you!
… oh, and go read my paper… please. Dieni, C.A., Bouffard, M.C., and Storey, K.B. (2012) Glycogen synthase kinase-3: cryoprotection and glycogen metabolism in the freeze-tolerant wood frog. J. Exp. Biol. 215: 543-551.
.
.
Chris is originally from Montreal, and is a Comparative Biochemist and Physiologist. His educational and postdoctoral experiences have taken him from Montreal to Ottawa ON, State College PA, and finally back to Montreal’s biotech industry. In his spare time- as you would expect from a Canadian- Chris enjoys watching hockey and is a stalwart fan of the Montreal Canadiens and Ottawa Senators. You can keep up to date with the latest from Chris on Twitter.
.
Have any kinase-related questions for Chris?
.


brian
wrote on April 20, 2012 at 2:31 pm
is phosphorylation the most common post translational signaling mechanism, versus nitrosylation or other modifications?
@chrisdieni
wrote on April 20, 2012 at 6:41 pm
Hey Brian, thanks for the question! From our current understanding of post-translational regulation, yes, protein phosphorylation is the most common mechanism. Approximately one-third of eukaryotic proteins are phosphorylated in any given physiological state (Zolnierowicz, S. and Bollen, M. (2000) Protein phosphorylation and protein phosphatases. EMBO. J. 19: 483-488.). A little over a year ago, a bioinformatics company in Vancouver, BC, Canada called Kinexus identified over 650,000 potential phosphorylation sites in the human proteome alone (http://www.prnewswire.com/news-releases/kinexus-identifies-over-650000-phosphorylation-sites-in-the-human-proteome-116722899.html). Comparatively speaking, these factors make phosphorylation far more prevalent than any other PTM.
Hope this helps!