Tetrahymena: Little Creature, Big Discoveries
by
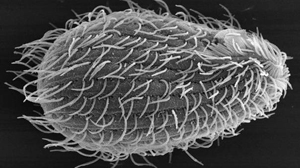 You probably didn’t notice, but if you’ve ever dipped a toe into a pond, or swam in a stream, you would have bumped into this furry looking microscopic creature called Tetrahymena. This fresh water inhabitant may be small and single celled, but it has played a huge role in discoveries that contributed to our understanding of the fundamental principles of eukaryotic biology.
You probably didn’t notice, but if you’ve ever dipped a toe into a pond, or swam in a stream, you would have bumped into this furry looking microscopic creature called Tetrahymena. This fresh water inhabitant may be small and single celled, but it has played a huge role in discoveries that contributed to our understanding of the fundamental principles of eukaryotic biology.
Tetrahymena belongs to an evolutionary lineage of unicellular eukaryotes – ciliates. It’s a tough little creature, and ecologically very successful. It can live in a wide range of temperatures and adapt to diverse environments. Although people have been aware of its existence since the 1800s, it was the Nobel laureate Andre Lwoff who successfully cultured this organism in 1923. Since then, Tetrahymena has become a prominent model organism and contributed to many pivotal findings in biology. Nobel prize winning discoveries of ribozymes and telomere function are just a few examples. With its somatic genome sequenced, today, Tetrahymena’s potential in aiding groundbreaking scientific discoveries seems endless.
Unicell, complex biology
Part of what makes Tetrahymena an excellent model organism is that it possesses the advantages of other laboratory single cell models, yet, it shares with us (multicellular animals) many genes and processes typically absent in single celled organisms. It’s pretty spectacular that this unicell takes nearly as many genes as humans do (~25,000). Although single celled, its genome and cellular functions make it look like more complex metazoa. What’s so cool? Where multicellular organisms divide complexity amongst different cells, this organism miraculously packs it into one.
Exactly how complex is Tetrahymena? And how can we take advantage of its quirky features to gain insights into the fundamental principles of biology? Here are a few examples:
Tetrahymena’s single cell exhibits extraordinary polarity. It has a mouth apparatus that it eats with, and a posterior end analogous to, well… It’s hundreds of cilia are perfectly aligned in a specific orientation, which is inherited epigenetically. What does that mean? If you take a cell, and invert the orientation of a row of cilia, its progeny will inherit the new pattern without any changes to its DNA sequences. These features make Tetrahymena a tool to study cellular patterning, axis formation and epigenetic inheritance.
Did you know that Tetrahymena’s sophisticated cytoskeletal network was behind the identification and purification of the first cytoskeletal motor, dynein?
Nervous system and digestive machineries are common in multicellar organisms, but who would have thought that Tetrahymena, being a single cell, would have them too? It possesses a diverse sensory system to seek out food, and an eating machine that allows it to digest just about any protein it encounters. Tetrahymena expresses genes that are homologs of many neurotransmitter receptors. It is quickly becoming a model organism for studying neurobiology. Needless to say, pharmaceutical companies are already using Tetrahymena for drug development and testing.
Like multicellular organisms, Tetrahymena maintains both germline and somatic genomes. How do you do that when you have only one cell? Tetrahymena organizes the two distinct genomes into separate nuclei. The smaller of the two nuclei is transcriptionally silenced and reserved for sex. It is analogous to our germline and carries the genetic information passed on to the sexual progeny. The bigger nucleus, also known as the soma, does not directly contribute to the genetic composition of the sexual progeny, but is responsible for producing all the gene products necessary to support vegetative growth (non-sexual stage of the life-cycle where cells divide by binary fission). During sex, the making of the new somatic nucleus involves genome wide DNA rearrangement to remove transposable elements and ‘junk’ DNA. The phenomena associated with nuclear dimorphism in Tetrahymena provide opportunities for us to study processes that are general to members across the animal kingdom. For example: how do the germline and the soma maintain and carry out their differential functions during the lifetime of an organism? What are the mechanisms that cells use to target transposons for silencing during the differentiation of the soma? What are the transcription dependent vs. independent mechanisms to regulate chromatin structures?
Advantages of using Tetrahymena for research:
Excellent surrogate for studying the functions of conserved proteins and processes.
Easy to maintain – You can easily freeze them for long-term storage; or keep them alive in a slow growing condition for 6 months without having to tend to them.
Short generation time – Tetrahymena is among the fastest growing eukaryotes. Its doubling time is below 2 hours and it can readily grow to high density. Sexual reproduction can be triggered at will and highly synchronized. The making of the sexual progeny takes less than 24 hours. What this means for graduate students, such as myself, is that we get to work all the time.
Cost efficient – All you need is a couple of flasks and/or petri dishes to house the cells. They aren’t picky eaters. Out in the wild, they probably gobble down just about anything they encounter. In the lab, they will reproducibly grow to high density with media containing Proteose Peptone and a source of iron supplement.
Availability of genetic and molecular tools – Homologous recombination allows targeted manipulation of specific genes. Knocking out a gene takes just a few days. You can easily tag pretty much any gene and drop it into its endogenous locus. If you want to go the ectopic route, or hook your gene up to inducible promoters, there are well established systems set up just for that. Basically, studying in vivo function of a gene couldn’t be easier.
Drawbacks
Gene knockdown with RNAi does not work very well in Tetrahymena.
Perhaps one of the biggest drawbacks of Tetrahymena is that it uses an alternate genetic code. Pretty much every living being on earth follows the universal genetic code. But in Tetrahymena, two of the stop codons used by all the other organisms actually encode amino acids. This makes it difficult to express Tetrahymena genes in other organisms. And the use of techniques developed in other systems (such as yeast-two hybrid screen) is less feasible.
_________________________________________________________
Annie Shieh is a graduate student of Developmental Biology at Washington University in St. Louis. She is studying programmed DNA rearrangement during Tetrahymena sexual reproduction. When she is not working in the lab, she enjoys traveling, dancing and biking in the park.
_________________________________________________________
Resources
Orias, E. (2002) Sequencing the Tetrahymena thermophila genome. White paper.
Tetrahymena genome database – Tetrahymena biology. http://www.ciliate.org/Biology.shtml
Asai, DJ. Forney, JD. (2000) Methods in Cell Biology – Tetrahymena thermophila. Academic press: USA
_________________________________________________________
See other articles in the series:
Drosophila melanogaster- The fruit fly.
Research’s Next Top Model (Zebrafish)
Getting to Know Your Worms (C. elegans)
Saccharomyces cerevisiae a.k.a Budding/Baker’s/Brewer’s Yeast
The Almighty Fungi: The Revolutionary Neurospora crassa
We’re Gonna Need a Bigger Lab: Large Animal Models in Research
.


guestUVM
wrote on February 22, 2010 at 2:40 am
RNAi sure seems to work in the related paramecium, would have expected it to work in tetrahymena. but great blog Ms Shieh! good work..
Drosophila melanogaster: The Fruit Fly | BenchFly Blog
wrote on May 24, 2010 at 11:07 am
[…] Tetrahymena: Little Creature, Big Discoveries […]
A Primer on Large Animal Models in Research | BenchFly Blog
wrote on May 24, 2010 at 11:11 am
[…] Tetrahymena: Little Creature, Big Discoveries […]
Saccharomyces cerevisiae: Yeast Rising in Research | BenchFly Blog
wrote on May 24, 2010 at 11:22 am
[…] Tetrahymena: Little Creature, Big Discoveries […]
Porcupine Paul
wrote on March 11, 2012 at 4:34 pm
Thank you so much! Really helpful :)
Jenni Santos
wrote on May 22, 2012 at 1:37 pm
Hi_I am a graduate student that is considering working with tetrahymena( i know nothing about it, have never grown protozoa) This was really helpful. any articles/ protocols or books you recomend? my email is [email protected]_Thanks!!